order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Location
Corporate Headquarters
Vector Laboratories, Inc.
6737 Mowry Ave
Newark, CA 94560
United States
Telephone Numbers
Customer Service: (800) 227-6666 / (650) 697-3600
Contact Us
Additional Contact Details
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
order histories, retained contact details for faster checkout, review submissions, and special promotions.
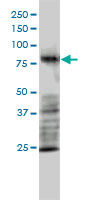
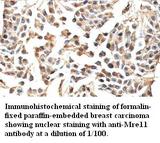
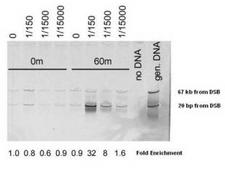
MRE11A / MRE11
MRE11 meiotic recombination 11 homolog A (S. cerevisiae)
Component of the MRN complex, which plays a central role in double-strand break (DSB) repair, DNA recombination, maintenance of telomere integrity and meiosis. The complex possesses single-strand endonuclease activity and double-strand-specific 3'-5' exonuclease activity, which are provided by MRE11A. RAD50 may be required to bind DNA ends and hold them in close proximity. This could facilitate searches for short or long regions of sequence homology in the recombining DNA templates, and may also stimulate the activity of DNA ligases and/or restrict the nuclease activity of MRE11A to prevent nucleolytic degradation past a given point. The complex may also be required for DNA damage signaling via activation of the ATM kinase. In telomeres the MRN complex may modulate t-loop formation.
| Gene Name: | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| Synonyms: | MRE11A, AT-like disease, Endo/exonuclease Mre11, HNGS1, MRE11, MRE11 homolog 1, MRE11 homolog A, MRE11B, ATLD |
| Target Sequences: | NM_005591 NP_005582.1 P49959 |
Publications (88)
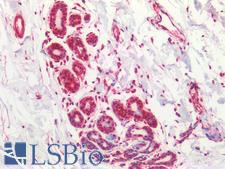

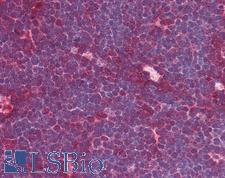
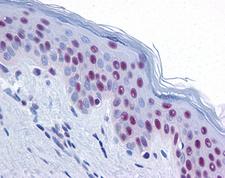
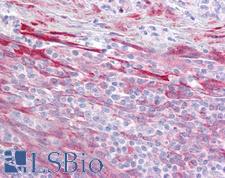
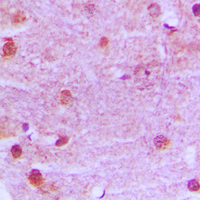
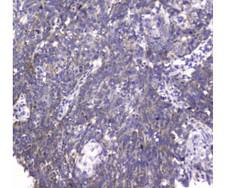
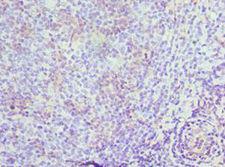
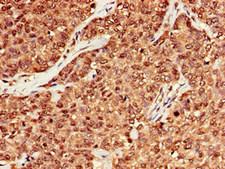

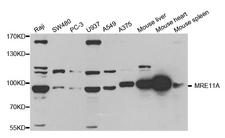
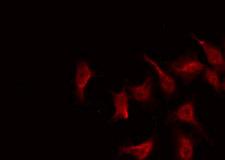

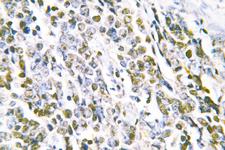
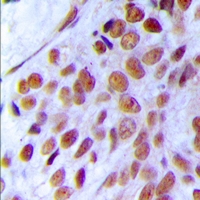
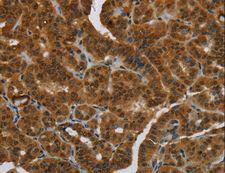
If you do not find the reagent or information you require, please contact Customer.Support@LSBio.com to inquire about additional products in development.













