order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Location
Corporate Headquarters
Vector Laboratories, Inc.
6737 Mowry Ave
Newark, CA 94560
United States
Telephone Numbers
Customer Service: (800) 227-6666 / (650) 697-3600
Contact Us
Additional Contact Details
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Forgot password?
order histories, retained contact details for faster checkout, review submissions, and special promotions.
Coronavirus Research Products
In support of the global research effort, we offer SARS-CoV-2 Antibodies, Detection Kits, Proteins, cDNA clones, Inhibitors, and Cytokine Release Syndrome research products.
All of our Coronavirus Research Products are for research use only.
Coronavirus Detection Kits
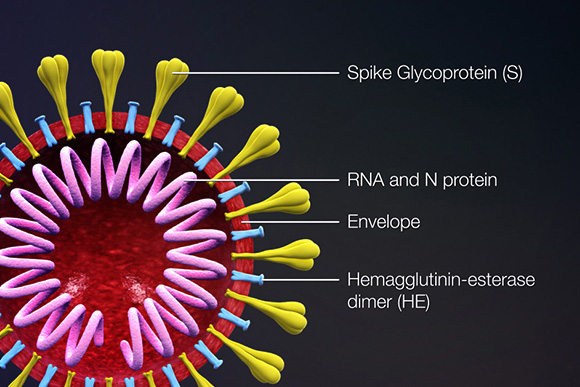
Coronavirus Antibodies, Proteins, and cDNAs
Coronavirus Inhibitors
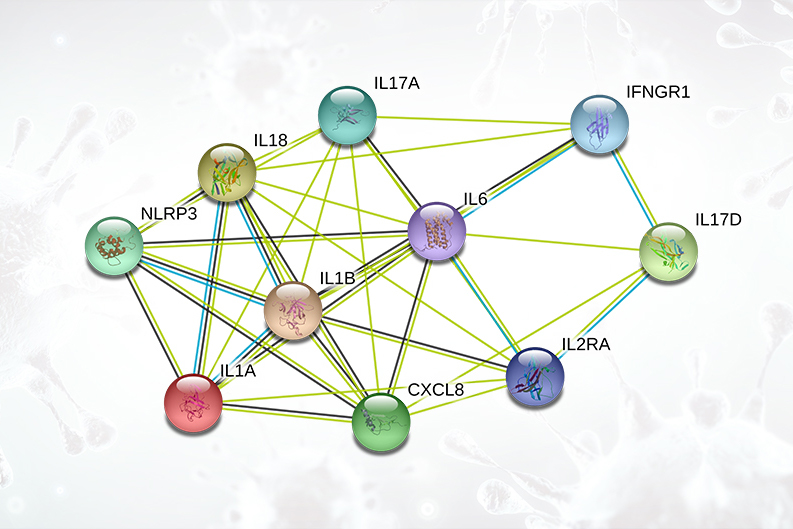
Cytokine Release Syndrome
Coronaviruses (CoVs) are enveloped viruses with positive-sense, single-stranded RNA genomes that range in size from 26 to 32 kilobases, the largest among RNA viruses. They are pathogens known to infect humans and numerous other animal species. Based on genetic and antigenic criteria, CoVs have been organized into four groups: alpha, beta, gamma, and delta coronaviruses (Dhama, 2014; Coleman, 2014).
Common human coronaviruses include the 229E and NL63 alpha coronaviruses and the OC43 and HKU-1 beta coronaviruses. In infected humans, they are associated with a range of cold-like symptoms as well as severe respiratory tract infections (Fielding, 2011). Other more symptomatically severe human coronaviruses that have been transmitted from animals include MERS-CoV, a beta coronavirus that causes Middle East Respiratory Syndrome (MERS); SARS-CoV, a beta coronavirus that causes severe acute respiratory syndrome (SARS); and SARS-CoV-2, a beta coronavirus that causes coronavirus disease 2019 (COVID-19).
Proteins that contribute to the overall structure and function of all coronaviruses are the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as the non-structural proteins cleaved from the orf1a/b polyprotein. Important non-structural proteins include the RNA-dependent RNA polymerase, protease, and endoRNAse (Fehr, 2016). The spike (S) protein is responsible for attachment to and fusion with the host cell. For both SARS-CoV and SARS-CoV-2, the host receptor is known to be Angiotensin converting enzyme 2 (ACE2) (Hoffman, 2020). The nucleocapsid (N) protein binds to the CoV genome and is responsible for packaging it into the ribonucleoprotein complex (capsid). The membrane (M) protein determines the shape of the viral envelope and organizes viral assembly through interaction with each additional structural protein. The envelope (E) protein is the smallest of the structural proteins and is incorporated into the virion envelope, but this represents only a small amount of total expressed envelope protein. A high proportion is also expressed inside the infected cell, where it is involved in viral assembly, budding, maturation and propagation (Schoeman, 2019; Chang, 2020).
The SARS-CoV-2 (COVID-19) coronavirus genome has 89% sequence similarity with the bat SARS-like CoVZXC21 coronavirus, and 82% similarity with human SARS-CoV coronavirus. The orf1a/b, S, E, M, and N proteins also share a high degree of phylogenetic similarity with bat, civet, and human SARS coronaviruses. Notable differences in sequence are found in the SARS-CoV-2 spike protein’s receptor binding domain, which has only 40% amino acid overlap with other related viruses. Furthermore, the SARS-CoV-2 orf3b protein represents a completely novel short protein (Rehman, 2020; Chan, 2020).
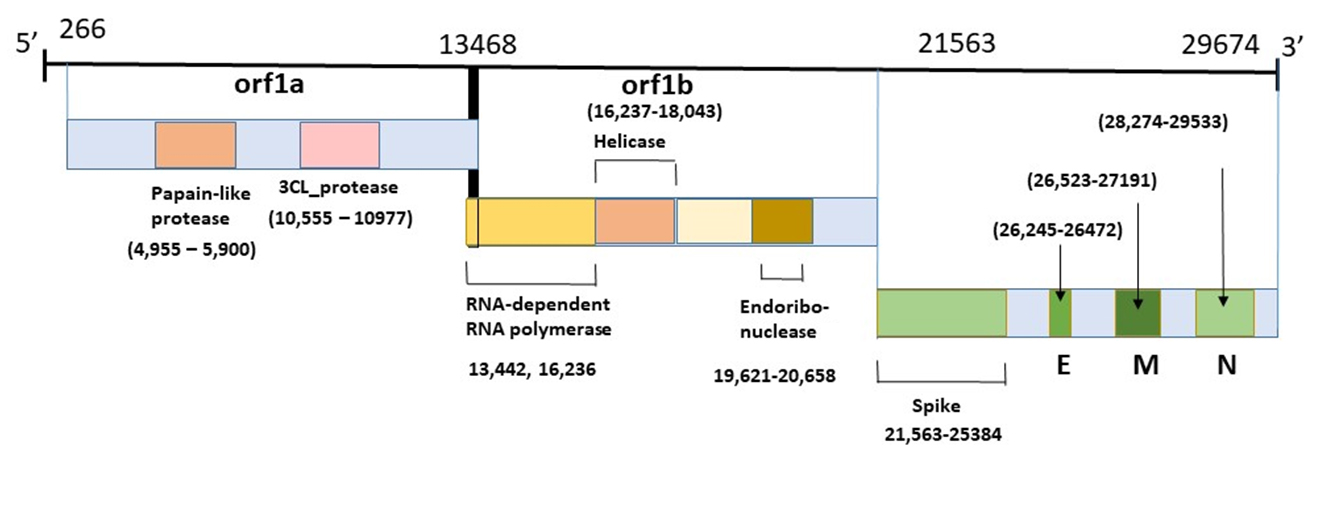
Coronavirus Genome
This diagram illustrates the genomic organization of the 29,674-nucleotides of the SARS-CoV-2 coronavirus. The regions that encode predicted ORFs 1a and 1b, which encode the nonstructural polyproteins (nsp1-16, helicase, 3CL-pro and PL-pro, RNA-dependent RNA polymerase), as well as the spike, envelope, membrane, and nucleocapsid structural proteins are indicated.
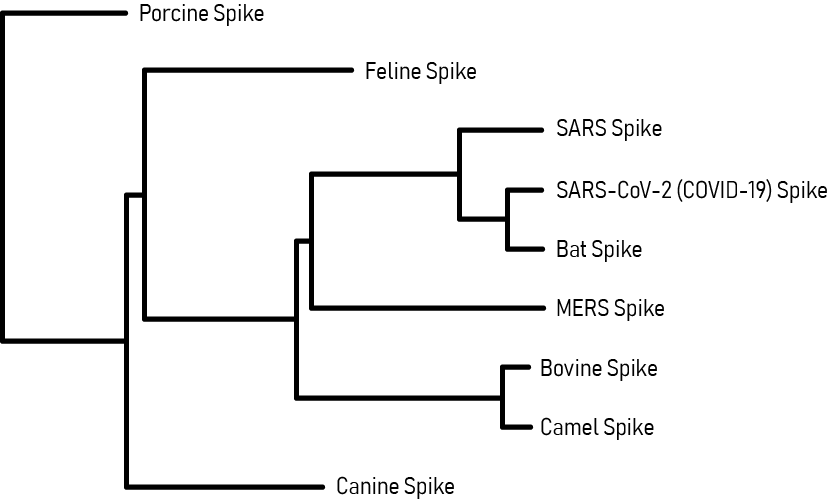
Coronavirus Spike Glycoprotein Phylogenetic Tree
This phylogenetic tree maps the evolutionary relationships of the SARS-CoV-2 Spike (S) glycoprotein with other coronaviruses. Analysis of the SARS-CoV-2 S protein indicated close proximity with the SARS-CoV and Bat RaTG13 coronaviruses, which are thought to be its closest relatives and potential origins. The next proximal relation is with the MERS coronavirus. Phylogenetic analysis is useful for coronavirus disease research and in the development of potential therapies, since drugs developed for closely related viruses (SARS-CoV, MERS) may be functionally repurposed as potential anti-SARS-CoV-2 agents.
Coronavirus Family Spike Glycoprotein Homology Heatmap
The heatmap below displays the degree of sequence similarity of the SARS-CoV-2 Spike glycoprotein with that of other coronaviruses including SARS-CoV and MERS. It demonstrates that SARS-CoV-2 spike protein has high homology with the Bat RaTG13 and SARS-CoV Spike proteins, moderate similarity with bovine spike protein, and relatively low homology with canine coronavirus spike proteins.
| FELINE | CANINE | PORCINE | SARS-CoV | SARS-CoV-2 (COVID-19) | BAT | MERS | BOVINE | CAMEL | |
|---|---|---|---|---|---|---|---|---|---|
| FELINE (ASU62488.1) | 100.00 | 49.53 | 42.42 | 28.64 | 28.48 | 28.66 | 26.54 | 29.11 | 29.2 |
| CANINE (AKA65829.1) | 49.53 | 100.00 | 41.95 | 25.5 | 25.76 | 25.76 | 25.72 | 25.2 | 25.11 |
| PORCINE (AIA61001.1) | 42.42 | 41.95 | 100.00 | 26.37 | 26.46 | 26.35 | 26.85 | 27.17 | 27.48 |
| SARS-CoV (NP_828851.1) | 28.64 | 25.5 | 26.37 | 100.00 | 77.77 | 78.23 | 32.16 | 32.73 | 32.56 |
| SARS-CoV-2 (YP_009724390.1) | 28.48 | 25.76 | 26.46 | 77.77 | 100.00 | 97.71 | 31.79 | 32.13 | 32.13 |
| BAT (QHR63300.2) | 28.66 | 25.76 | 26.35 | 78.23 | 97.71 | 100.00 | 31.79 | 32.08 | 32.25 |
| MERS (YP_009047204.1) | 26.54 | 25.72 | 26.85 | 32.16 | 31.79 | 31.79 | 100.00 | 34.21 | 34.54 |
| BOVINE (NP_150077.1) | 29.11 | 25.20 | 27.17 | 32.73 | 32.13 | 32.08 | 34.21 | 100.00 | 94.28 |
| CAMEL (AHN64774.1) | 29.20 | 25.11 | 27.48 | 32.56 | 32.13 | 32.25 | 34.54 | 94.28 | 100.00 |
| Key |
|---|
| 100.00 |
| 83.33 |
| 66.66 |
| 50.00 |
| 33.33 |
| 16.66 |
| 0.00 |
References
-
Chan et al. Coronavirus genome similarity: Emerg Microbes Infect. 2020 Jan 28;9(1):221-236. doi: 10.1080/22221751.2020.1719902
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7067204/ -
Chang et al. Anti-viral Research Volume 103, March 2014, Pages 39-50
https://www.sciencedirect.com/science/article/pii/S0166354213003781?via%3Dihub - Coleman, CM, Frieman, MB. J Virol. 2014 May;88(10):5209-12. doi: 10.1128/JVI.03488-13
- Dhama, K., Pawaiya, R.V.S., Chakraborty, S., Tiwari,R., Saminathan, M., and A.K. Verma, 2014. Coronavirus Infection in Equines: A Review. Asian Journal of Animal and Veterinary Advances, 9: 164-176.
-
Fehr, A, Perlman, S. Methods Mol Biol. 2015;1282:1-23. doi: 10.1007/978-1-4939-2438-7_1
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4369385/ -
Fielding, B. Future Microbiol. 2011 Feb; 6(2): 153–159. DOI: 10.2217/fmb.10.166
-
Hoffmann et al. SARS-CoV-2 ACE2-dependent entry: Hoffmann et al. 2020, Cell 181: 1–10. DOI: 10.1016/j.cell.2020.02.052
https://www.cell.com/cell/pdf/S0092-8674(20)30229-4.pdf -
Rehman et al. Pathogens 2020, 9(3), 240. DOI: 10.3390/pathogens9030240
https://www.mdpi.com/2076-0817/9/3/240/htm -
Schoeman, D, Fielding, B. Virology Journal volume 16, Article number: 69 (2019)
https://virologyj.biomedcentral.com/articles/10.1186/s12985-019-1182-0










